Pokemon Statistics
library(tidyverse)## Warning: package 'dplyr' was built under R version 4.2.3## Warning: package 'stringr' was built under R version 4.2.3## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.4
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.4.4 ✔ tibble 3.2.1
## ✔ lubridate 1.9.3 ✔ tidyr 1.3.0
## ✔ purrr 1.0.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(glmnet)## Loading required package: Matrix## Warning: package 'Matrix' was built under R version 4.2.3##
## Attaching package: 'Matrix'
##
## The following objects are masked from 'package:tidyr':
##
## expand, pack, unpack
##
## Loaded glmnet 4.1-8set.seed(11)bwt_df =
read_csv("./birthweight.csv") %>%
janitor::clean_names() %>%
mutate(
babysex = as.factor(babysex),
babysex = fct_recode(babysex, "male" = "1", "female" = "2"),
frace = as.factor(frace),
frace = fct_recode(frace, "white" = "1", "black" = "2", "asian" = "3",
"puerto rican" = "4", "other" = "8"),
malform = as.logical(malform),
mrace = as.factor(mrace),
mrace = fct_recode(mrace, "white" = "1", "black" = "2", "asian" = "3",
"puerto rican" = "4")) %>%
sample_n(200)## Rows: 4342 Columns: 20
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (20): babysex, bhead, blength, bwt, delwt, fincome, frace, gaweeks, malf...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Inputs fir “glmnet”
x = model.matrix(bwt ~ ., bwt_df)[,-1]#[,-1] no intercept
y = bwt_df$bwtFit Lasso!
lambda = 10^(seq(3, -2, -0.1)) # illustare how lasso work
lasso_fit =
glmnet(x, y, lambda = lambda)
lasso_cv =
cv.glmnet(x, y, lambda = lambda) # best lamda
lambda_opt = lasso_cv$lambda.minThis is the plot for lasso
broom::tidy(lasso_fit) %>%
select(term, lambda, estimate) %>%
complete(term, lambda, fill = list(estimate = 0) ) %>%
filter(term != "(Intercept)") %>%
ggplot(aes(x = log(lambda, 10), y = estimate, group = term, color = term)) +
geom_path() +
geom_vline(xintercept = log(lambda_opt, 10), color = "blue", size = 1.2) +
theme(legend.position = "none")## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.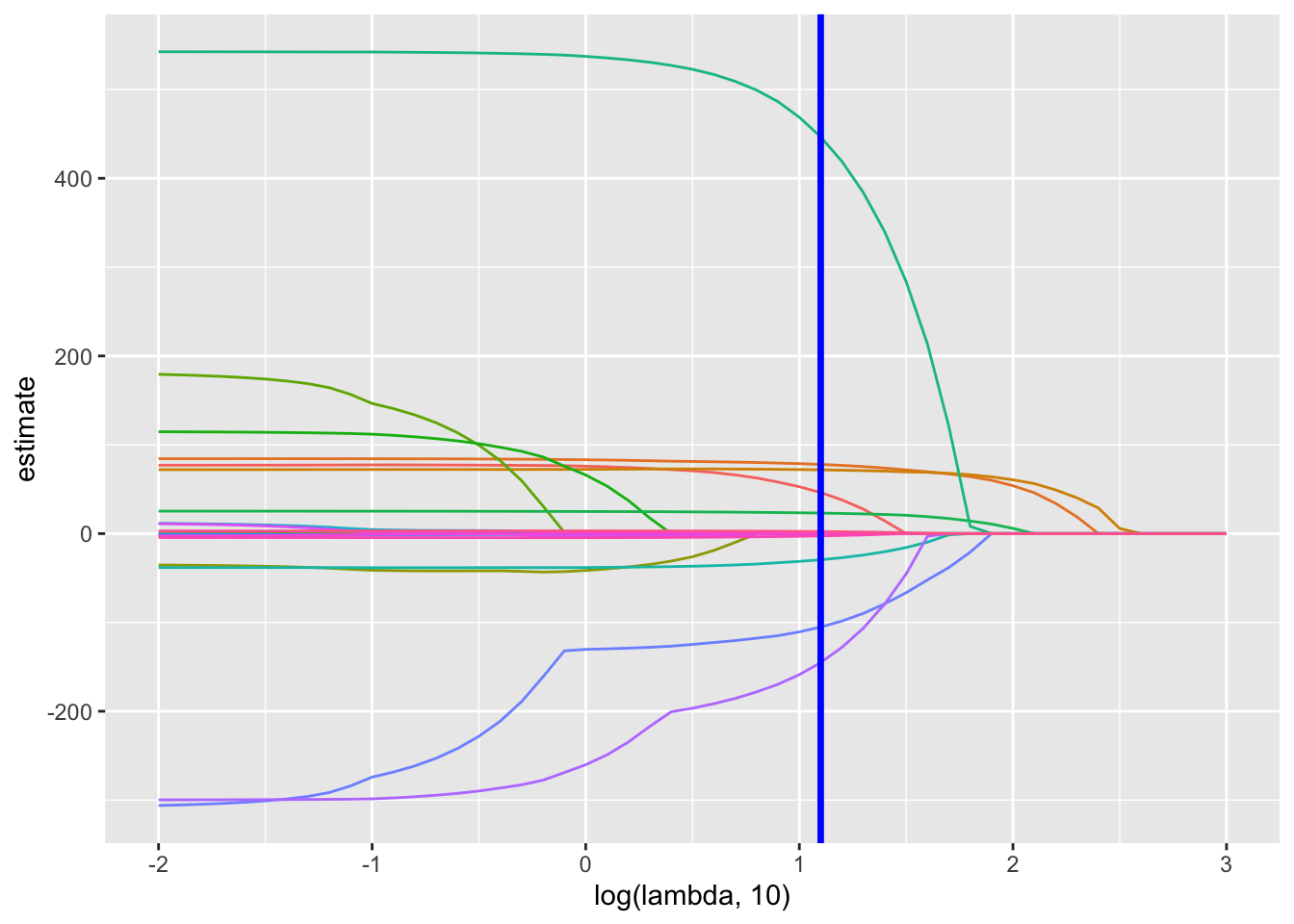
broom::tidy(lasso_cv) %>%
ggplot(aes(x = log(lambda, 10), y = estimate)) +
geom_point() 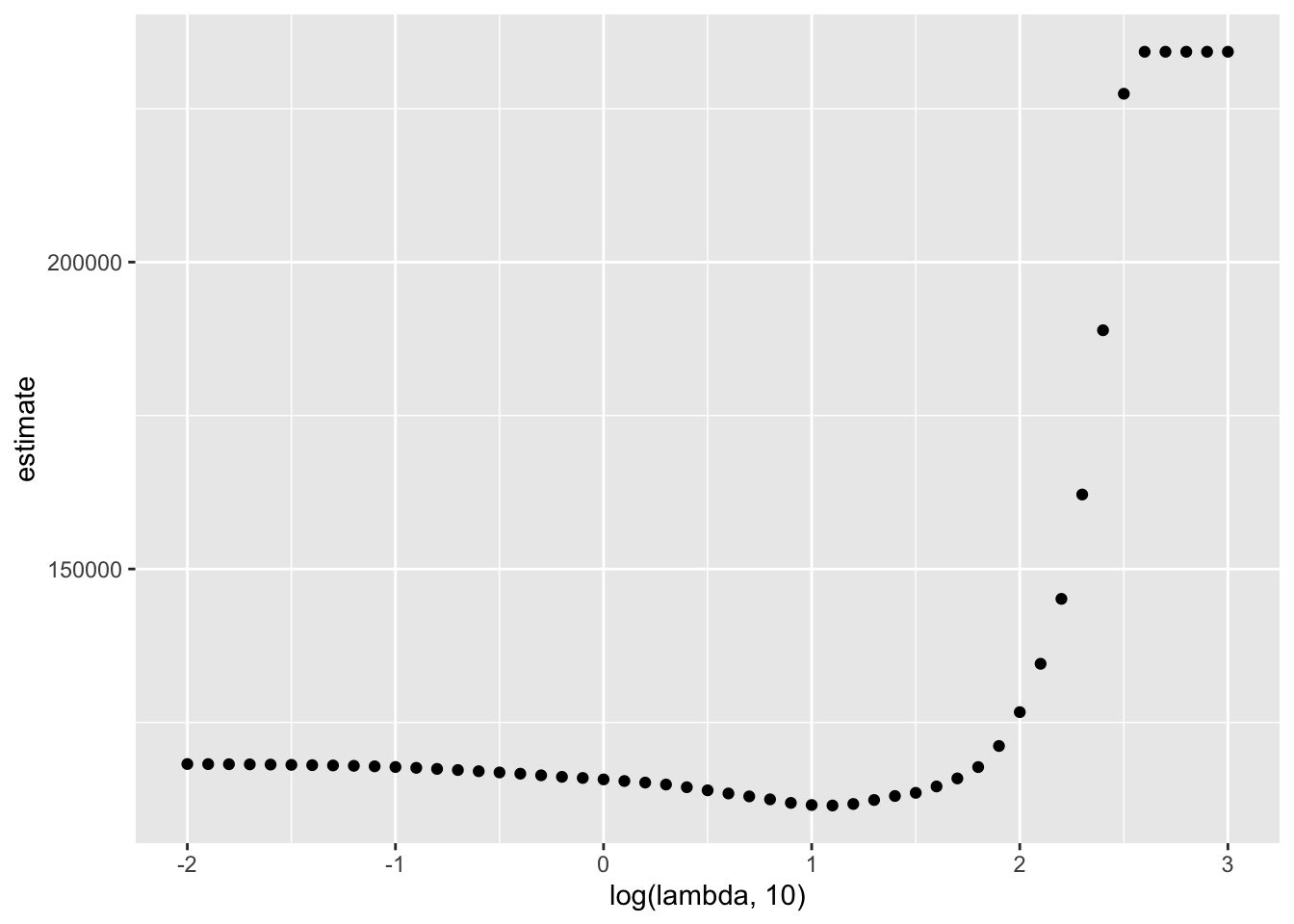
Clustering
poke_df =
read_csv("./pokemon.csv") %>%
janitor::clean_names() %>%
select(hp, speed)## Rows: 800 Columns: 13
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): Name, Type 1, Type 2
## dbl (9): #, Total, HP, Attack, Defense, Sp. Atk, Sp. Def, Speed, Generation
## lgl (1): Legendary
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.poke_df %>%
ggplot(aes(x = hp, y = speed)) +
geom_point()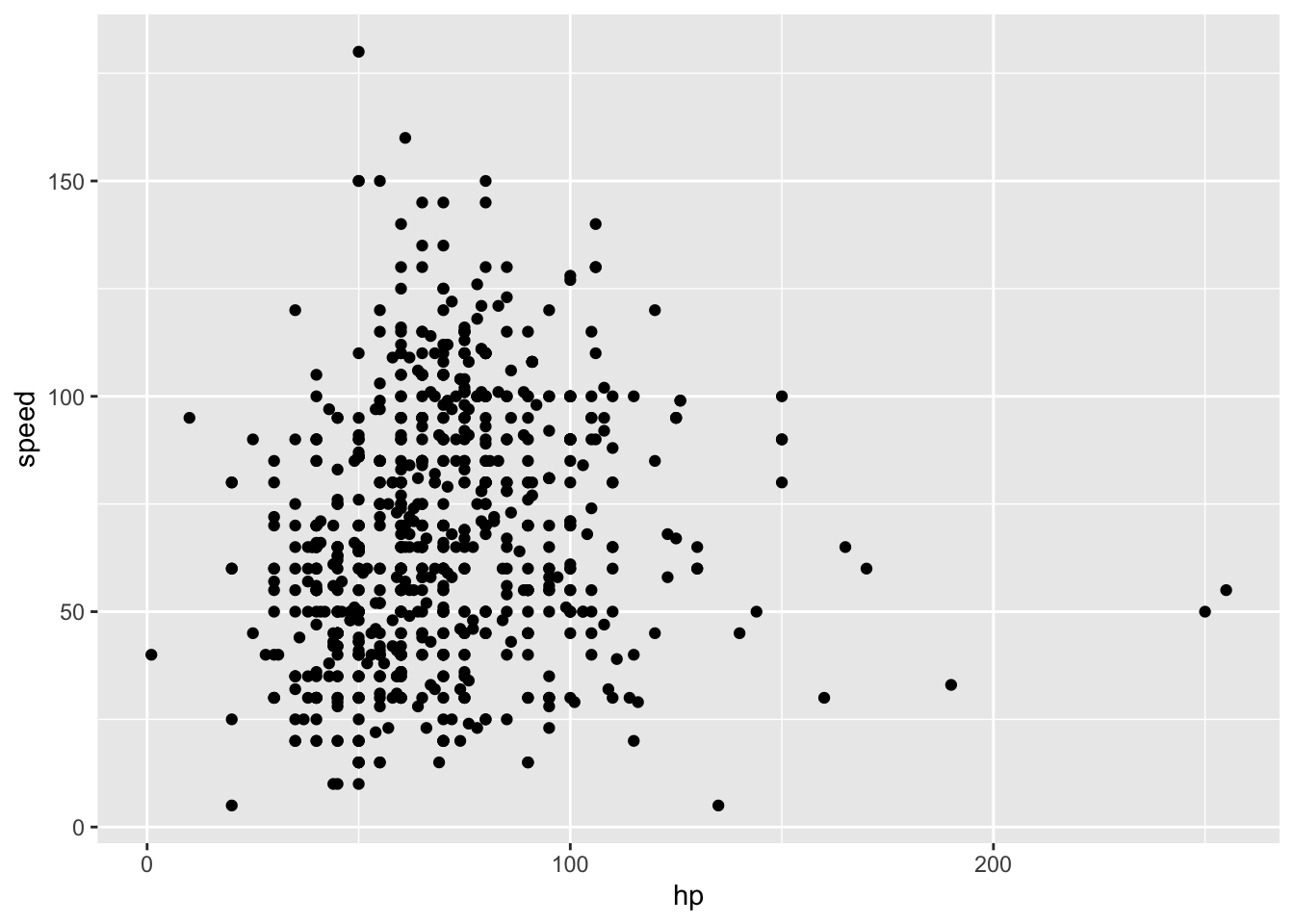 Run K means
Run K means
kmeans_fit =
kmeans(x = poke_df, centers = 3) # run cluster and want 3 clusterspoke_df =
broom::augment(kmeans_fit, poke_df) # give me cluster mean, imorted cluste to my original dataset
poke_df %>%
ggplot(aes(x = hp, y = speed, color = .cluster)) +
geom_point()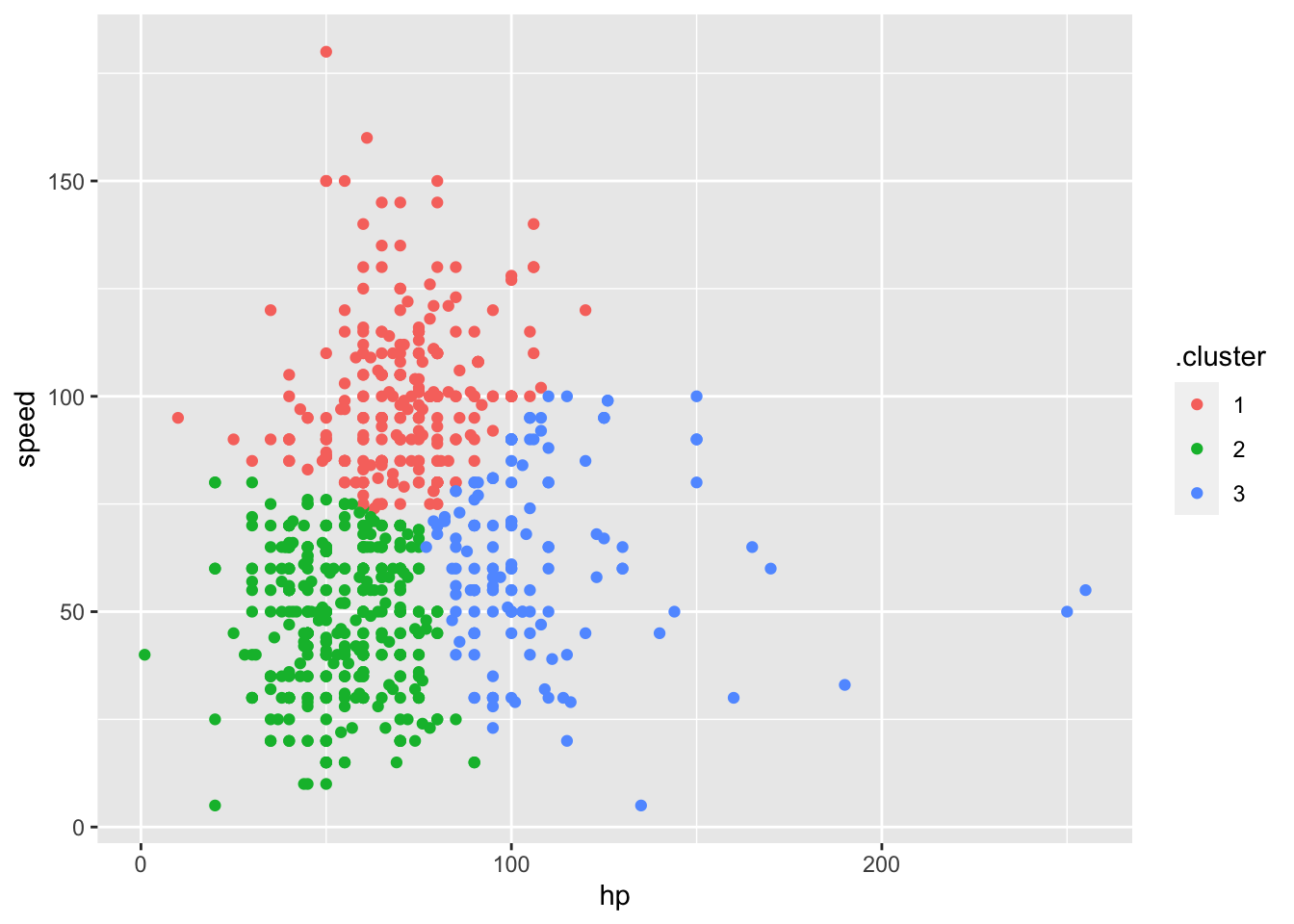
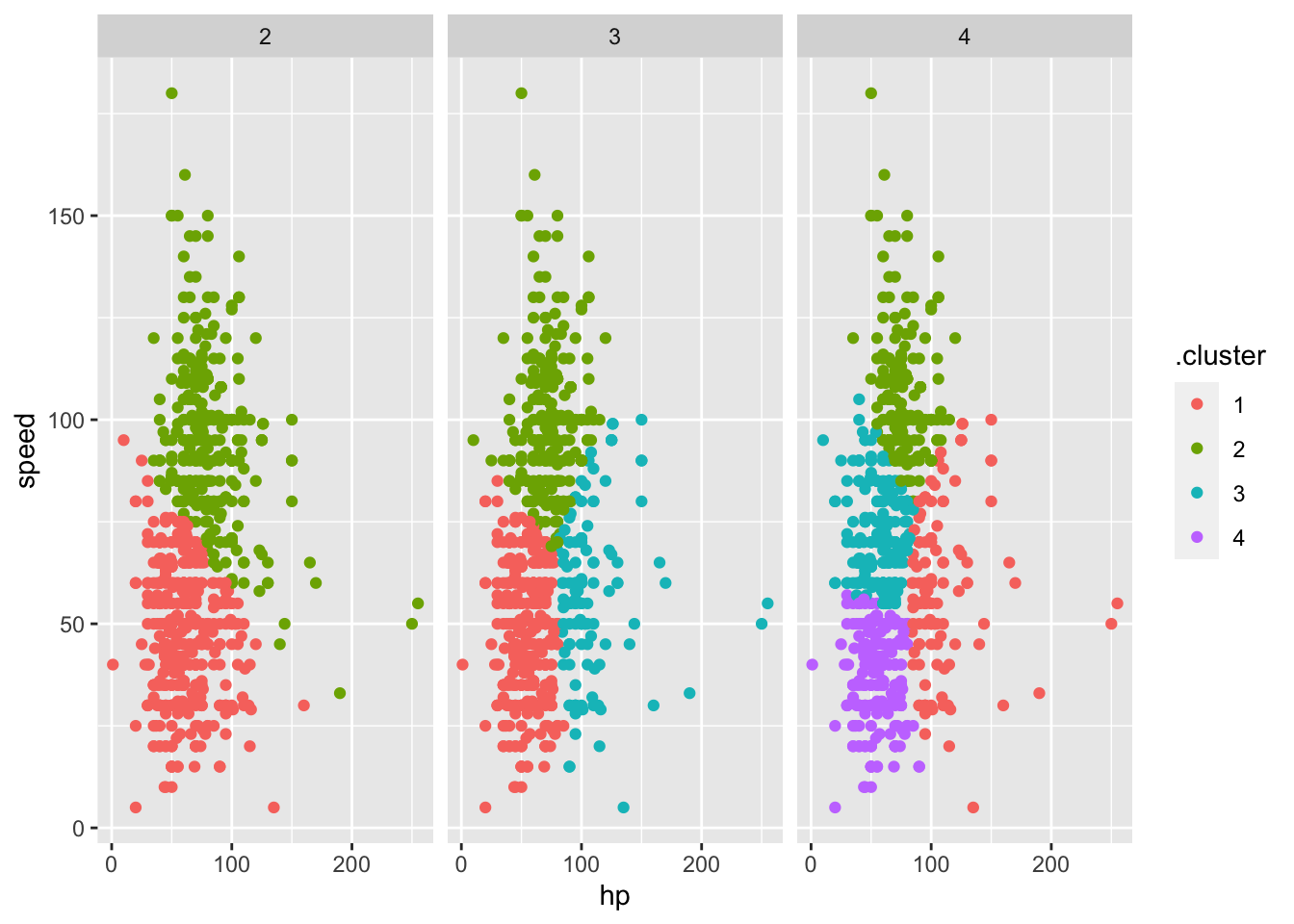
clusts =
tibble(k = 2:4) %>%
mutate(
km_fit = map(k, ~kmeans(poke_df, .x)),
augmented = map(km_fit, ~broom::augment(.x, poke_df))
)
clusts %>%
select(-km_fit) %>%
unnest(augmented) %>%
ggplot(aes(hp, speed, color = .cluster)) +
geom_point(aes(color = .cluster)) +
facet_grid(~k)